Target-based drug discovery (TBDD) strategies followed by pharma companies involve understanding the disease’s mechanism of action, followed by target identification and validation.
This process allows scientists and researchers to identify and work with a specific gene, gene product or molecular mechanism as responsible for the disease under investigation. Recombinant technology will offer screening against large compound libraries to identify “hits” – small molecules and/or biologics that interact with the target of interest. Further improvements of the selected hits through a series of steps, such as hit-to-lead and lead optimization, will identify and select a final candidate molecule which will progress towards clinical trials.
In contrast, the other strategy for drug discovery is phenotypic or target-agnostic screening where drugs are identified without prior knowledge of a specific molecular target. Instead, drugs are tested for their pharmacological actions. Antibiotics are a good example of medicines discovered in this way.
With rapid advances in molecular biology, genomics, and proteomics, TBDD has become the leading approach adopted by the pharmaceutical industry for the past three decades. A main advantage of TBDD is its ease of execution compared to the phenotypic approach. It is also generally faster and less costly.
Technological advances in the field of computer science have given rise to machine learning and AI-assisted models that can drastically improve the outcomes of TBDD through virtual drug screening and de novo drug design.
Potential opportunities with TBDD
The cosmetic sector has not yet sufficiently exploited such technologies and developments and it can be argued that TBDD could represent a new source of promising active molecules, backed by a clear scientific rationale increasing the value of those compounds.
In Silico Discovery (ISD) was founded with the objective of developing a target-based computational approach for the efficient discovery of new potential actives with application in the cosmeceutical area.
ISD achieves this through a solid technological partnership with Nostrum BioDiscovery (NBD), a highly specialized company providing custom-made services and cutting-edge in silico technologies to biopharmaceutical and biotechnology companies, CROs and academia to perform rational drug design and protein engineering.
Target cell population
The epidermis, which consists primarily of keratinocytes, is continuously renewed by the proliferation of stem cells and the differentiation of their progeny. Stem cells are then instrumental in epidermal renewal, regeneration, and repair. Different pools of stem cells are located in different epidermal regions, including the hair follicle, the interfollicular epidermis, and the sebaceous glands. Interfollicular epidermal stem cells (IFE-SCs) in the stratum basale play an especially important role in epidermal homeostasis under normal conditions and this population has been described to decrease during aging.
Given the relevant role of these cells in skin homeostasis, ISD has designed a first portfolio of targets able to modulate the behaviour of these cells and improve primarily their proliferation and barrier functions.
Target selection
In the selection of ISD’s targets, scientific publications related to cell population of interest and the availability of a 3D structure based primarily in X-ray or Cryo-EM were explored.
Today it is also possible to use AlphaFold, an AI model developed by DeepMind that predicts a protein’s 3D structure from its amino acid sequence. This breakthrough AI tool provides access to over 200 million protein structure predictions that are already accelerating scientific research.
Keeping a transversal point of view in the target selection process is critical as other scientific areas and therapeutic approaches could be of interest for the cosmetic sector. Topics such as senescence, autophagy, or mitochondria health, which are of interest in skin aging, are being pursued in oncology, diabetes or neurodegenerative therapeutic areas.
Different strategies to tackle targets
ISD has access to a plethora of proprietary algorithms (PELEplat, pyDock and ED/MD Suite Simulation) and third-party algorithms and computational technologies to challenge targets. The strategy to approach selected proteins is decided in the initial stages of each individual project. Two examples are detailed below.
One approach is to mimic the function of a protein of interest overcoming critical limitations such as a challenging recombinant synthesis because of its large size, structural complexity or low stability in solution. In these cases, it is possible to computationally build shorter versions of the protein and measure their in-silico stability and substrate binding properties, so that it keeps its original functionality.
In targets where it is possible to identify a potential binding site where a novel molecule may exert positive effects in the modulation of the activity of that protein and the consequent cellular signalling pathways, ISD will choose to run a high-throughput virtual screening using unique chemical or peptides libraries available at NBD. This strategy will follow the classical drug discovery “rocket”, where different computational tools will be applied to finally identify the best molecules.
These methods include molecular docking and molecular dynamics, among others, where the binding of a molecule towards the interesting target is estimated. The accuracy of such methods is gradually increased as the selection of compounds proceeds. This allows us to maintain a proper balance of computational cost – accuracy.
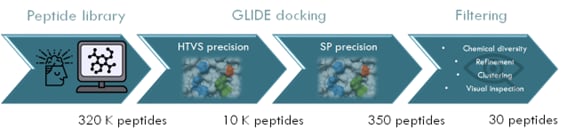
Figure 1: Example of a high-throughput virtual screening (HTVS) pipeline to discover active peptides. Glide is a molecular docking software that can perform binding predictions with different accuracy such as HTVS and SP.
High performance computing resources are frequently required to run such simulations, which can take several weeks of computation. ISD not only possesses an experienced team of computational scientists, but also the correct infrastructure to run such heavy calculations.
Conclusions
This in-silico approach focuses on a specific target, frequently used in pharmaceutical companies. It offers a highly efficient method in early stages of active ingredients identification, speeding up discovery and avoiding the use of precious experimental resources, which can be focused on key experiments. Moreover, this methodology provides a strong scientific hypothesis about the mechanism of action for these new cosmetic actives.
References
1. Swinney, D. C. (2013). Phenotypic vs. Target-Based Drug Discovery for First-in-Class Medicines. Clinical Pharmacology & Therapeutics, 93(4), 299-301. https://doi.org/10.1038/clpt.2012.236
2. Vincent, F., Nueda, A., Lee, J. et al. Phenotypic drug discovery: recent successes, lessons learned and new directions. Nat Rev Drug Discov 21, 899–914 (2022). https://doi.org/10.1038/s41573-022-00472-w
3. Iriyama, S., Yasuda, M., Nishikawa, S. et al. Decrease of laminin-511 in the basement membrane due to photoaging reduces epidermal stem/progenitor cells. Sci Rep 10, 12592 (2020). https://doi.org/10.1038/s41598-020-69558-y
4. Jumper, J., Evans, R., Pritzel, A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). https://doi.org/10.1038/s41586-021-03819-2


